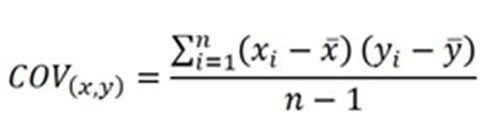A guide to Principal Component Analysis (PCA)
This article includes Principal Component Analysis working process.This concept is explained with examples and implementation using python.
- Introduction
- What is Principal Component Analysis (PCA)?
- Dimensionality reduction
- Why is it necessary to reduce data dimensions?
- How to perform dimensionality reduction using PCA
- Step-wise implementation of PCA
- Working process of PCA
- Example of PCA
- PCA implementation
- Conclusion
Introduction
Principal Component Analysis is one of the most sought-after machine learning techniques (PCA). The usefulness of PCA is its beauty. An essential outcome of applied linear algebra has been principal component analysis (PCA). Because PCA is an easy, non-parametric way of removing pertinent information from complex data sets, it is widely utilized in various studies, from neuroscience to computer graphics.



Best-suited Data Science courses for you
Learn Data Science with these high-rated online courses
Dimensionality reduction
It is important to understand the concept of dimensionality reduction to implement PCA effectively. Machine learning models are often susceptible to the dimensionality jinx. However, an efficient solution to this issue may be found with the help of dimensionality reduction. Dimensionality in statistics and machine learning refers to the number of features within a dataset. Dimensionality reduction is a strategy used to turn a high-dimensional space into a low-dimensional space by lowering the number of undesirable features within a dataset. The analysis of our feature space and selection of a subset of relevant features from this dataset is required following the dimensionality reduction. Future modeling then takes advantage of this subset.
There are various methods for dimensionality reduction, some of which include:
- Principal Component Analysis
- Linear Discriminant Analysis (LDA)
- Kernel PCA
- Canonical Correlation Analysis (CCA)
While detailing high dimensional data which are linearly separable, PCA is one of the most common techniques for dimensionality reduction.
Why is it necessary to reduce data dimensions?
- To avoid overfitting – as discussed above, the lesser assumptions a model makes, the simpler it will be.
- To reduce storage requirements – data with fewer features require lesser storage space.
- To speed up computation – data with fewer features train quicker.
- Removing redundant features and noise from the data to improve model performance can vastly increase model accuracy.
How to Perform Dimensionality Reduction Using PCA?
In large datasets, many features are often correlated. In other words, higher-dimensional data is dominated by a relatively small number of features. PCA is a method that linearly combines these highly correlated variables to form a smaller number of super features called the principal components. The components have the highest variance in the data. As an added benefit, the components, aka the PCs, will also be independent.
In the example below, the dataset has two features: x and y. All the data points are plotted as shown. PCA projects these data points in the direction of increasing variation. Let’s see how:
- It accomplishes this by identifying a set of new axes, or principal components, that are orthogonal (perpendicular) to each other in the direction of maximum variances to represent the data – where PC1 is the first axis that explains the largest variance, PC2 or the second axis accounts for the second-highest variance, and so forth.
- Then, each data point is projected onto the principal components with the maximum variance. In the above example, since the variance in the direction of PC2 is very less, data around PC2 is projected along the PC1 axis. The sum of variances pre and post-PCA is kept equal to ensure no data is lost.
The principal components can be equal to or less than the total number of data features. In mathematical terms, the principal components are the eigenvectors of the covariance matrix. Let’s look at the steps to implement PCA mathematically:
- Step1: Standardize the continuous features in the data.
This is done by subtracting the mean from each data point. This ensures that each attribute column has a mean of zero.
Standardizing or normalizing the features is important because each feature can be on a different scale and contribute significantly towards variance.
- Step2: Compute its covariance matrix.
For reference, the formula for covariance is given below:
The covariance matrix is given by:
- Step3: Compute the eigenvalues and eigenvectors for the above covariance matrix.
- Step 4: Sort the eigenvalues in descending order and select the largest n-eigenvalues, where n will be the number of axes required.
- Step 5: Select the respective eigenvectors that correspond to the eigenvalues mentioned above.
- Step 6: Multiply the original data with eigenvectors, which suggest the direction of new axes. Decide the features to keep based on which set accounts for 95% or more variance – these will be your PCs.
Step-wise implementation of PCA
Principal Component Analysis (PCA) is an important algorithm researchers and others use. By mapping a d-dimensional feature space onto a k-dimensional subspace, where k is lesser than d, the PCA method, a dimensionality reduction approach, decreases the dimension of a dataset.
By projecting all dependent features onto a new feature that is built in a method that minimizes the projection error, the PCA builds new features from the old ones.
This approach will ensure reliable findings if the data can be linearly separated.
The PCA technique has many applications, including feature extraction, gene analysis, and stock market forecasting.
The following stages are involved in PCA:
- Data standardisation.
- Obtaining the Eigen values and Eigen vectors with the help of the covariance or correlation matrix.
- The eigenvalues should be sorted in descending order.
- Select the first k eigenvectors, where k is the size of the new features space, and it is such that $(k<d)$ corresponds to the k’s biggest eigenvalues.
- Creating a projection matrix M using the chosen k eigenvectors.
- To create the new k-dimensional feature subspace Y, transform the original dataset X through M.



Working process of PCA
Principal Component Analysis seeks to reduce noise and redundancy by applying linear modifications to the data. The result is a dataset that highlights the most crucial aspects of the data. After using PCA, you will get a collection of principal components, and they will be ranked in decreasing order of how much they contribute to explaining patterns in the data. They are statistically evaluated based on how much variation they can explain. The first main component is crucial for characterizing data variance. The other primary components are less important for illustrating the variability of patterns in the data. Principal Component Analysis detects noise and redundancy in the dataset using statistical methods. The covariance matrix is used to examine the variation of each attribute. We may interpret that information in the diagonal cells to determine if a feature is significant or just random noise. Continuity and strength of the linear link between two characteristics. This is read in for all non-diagonal values and aids in identifying duplicate features.
After using PCA, you will get a collection of principal components, and they will be ranked in decreasing order of how much they contribute to explaining patterns in the data. They are statistically evaluated based on how much variation they can explain.
The first main component is crucial for characterizing data variance. The other primary components are less important for illustrating the variability of patterns in the data.
Principal Component Analysis detects noise and redundancy in the dataset using statistical methods. The covariance matrix is used to examine the variation of each attribute. We may interpret that information in the diagonal cells to determine if a feature is significant or just random noise. Continuity and strength of the linear link between two characteristics. This helps for all non-diagonal values and aids in identifying duplicate features.
The covariance matrix draws attention to the factors that make it difficult to detect patterns in the data. So, ultimately, PCA will result in a collection of primary components that:
- By boosting feature variance, reduce noise.
- By reducing the covariance between feature pairs, redundancy is reduced.
The covariance matrix serves as the foundation for PCA, and in actuality, there are two methods for determining the main components:
- Determine the covariance matrix’s eigenvectors.
- Calculate the covariance matrix’s Singular Value Decomposition.
Example of PCA
To demonstrate how PCA works and how it can be used to encode a smaller dataset, we will take the example of a real estate agent who is trying to determine why certain listings are closing too slowly. For instance, my friend X, a real estate agent, needs clarification as to why some of the properties managed by her business haven sold in more than six months. She would like to know if you could provide her with insight into what’s going on since she knows that you are a data scientist. The first step is to do some exploratory analysis. It is time to search for patterns in the data after doing a sanity check to determine if there are any missing or mismatched values. She is curious as to why these houses are sold later. She is interested in what aspects and trends in the data explain why these homes take so long to sell.
However, manually identifying patterns in a large dataset with many properties, also known as features, could be more practical.
Additionally, your collection can include many duplicated characteristics. Some characteristics are tiny changes of other features, you might think. Additionally, some characteristics can be random noises that don’t disclose anything about the fundamental patterns in the data.
For instance, you see that the following characteristics are present in her dataset:
- Total number of bedrooms,
- The total number of bathrooms, and
- The total number of doors in a home.
Since we already have the other two characteristics, having a feature for the total number of doors doesn’t have the potential to tell anything significant about the data. In terms of statistics, the total number of doors in a home is an unnecessary element and does not account for the variation in the data. No big thing, then. We may exclude the total number of doors in a home from the dataset if it is a redundant characteristic. This makes sense in our situation, but we should exercise more caution with a dataset with hundreds or thousands of characteristics. We must be certain that a feature is redundant or noisy before deleting it from a dataset containing many features. We need to realize it to delete crucial data points.
PCA implementation
The step-wise implementation process for PCA in Python is as follows:-
Import libraries
import numpy as npimport matplotlib.pyplot as pltimport numpy.random as rnd
Random data creation
# Create random 2d datamu = np.array([10,13])sigma = np.array([[3.5, -1.8], [-1.8,3.5]])print("Mu ", mu.shape)print("Sigma ", sigma.shape)# Create 1000 samples using mean and sigmaorg_data = rnd.multivariate_normal(mu, sigma, size=(1000))print("Data shape ", org_data.shape)
Data normalization
# Subtract mean from datamean = np.mean(org_data, axis= 0)print("Mean ", mean.shape)mean_data = org_data - meanprint("Data after subtracting mean ", org_data.shape, "\n")
Computation of variance matrix
# Subtract mean from datamean = np.mean(org_data, axis= 0)print("Mean ", mean.shape)mean_data = org_data - meanprint("Data after subtracting mean ", org_data.shape, "\n")
Eigen vectors’ computation for the covariance matrix along with sorting
# Perform eigen decomposition of covariance matrixeig_val, eig_vec = np.linalg.eig(cov)print("Eigen vectors ", eig_vec)print("Eigen values ", eig_val, "\n")# Perform eigen decomposition of covariance matrixeig_val, eig_vec = np.linalg.eig(cov)print("Eigen vectors ", eig_vec)print("Eigen values ", eig_val, "\n")
Computation of the explained variance and selecting N components
# Get explained variancesum_eig_val = np.sum(eig_val)explained_variance = eig_val/ sum_eig_valprint(explained_variance)cumulative_variance = np.cumsum(explained_variance)print(cumulative_variance)
Data transformation using eigen vectors
# Take transpose of eigen vectors with datapca_data = np.dot(mean_data, eig_vec)print("Transformed data ", pca_data.shape)
PCA inversion and computation of reconstruction data loss
# Compute reconstruction lossloss = np.mean(np.square(recon_data - org_data))print("Reconstruction loss ", loss)
Conclusion
The importance of PCA is increasing day by day. Various tools and software are being widely used for implementing it. Reducing the dimensionality of data with a linear distribution may be done effectively using PCA. Using PCA for picture data compression, it is possible to store large amounts of imaging data effectively, with lower dimensions, and without losing generality. However, previous knowledge of the data form is crucial to achieving a satisfactory PCA result in most situations. PCA cannot achieve effective data reduction when the provided data set has a nonlinear or multimodal distribution. Researchers have suggested dimension reduction approaches as PCA extensions, such as kernel PCA, multilinear PCA, and independent component analysis, to integrate the previous knowledge of the data into PCA.
Contributed by Sarthak
This is a collection of insightful articles from domain experts in the fields of Cloud Computing, DevOps, AWS, Data Science, Machine Learning, AI, and Natural Language Processing. The range of topics caters to upski... Read Full Bio