Feature selection Techniques|python code
Feature selection is a method of filtering out the important features as all the features present in the dataset are not equally important. There are some features that have no effect on the output. So we can skip them. As our motive is to reduce the data before feeding it to the training model. So feature selection is performed before training machine learning models. The models with less number of features have higher explainability, it is easier to implement machine learning models with reduced features. Feature selection removes data redundancy. Feature selection is performed after feature engineering.
This blog is mainly for those who want to learn feature selection implementation. But if you are not aware of the basics of feature selection or are a beginner then you can go and see my previous blog, where I covered feature selection techniques also and their differences. You don’t have to learn all the techniques. I will be covering the main important techniques for feature selection with their implementation. All these techniques are trying to find the common thing i.e dependency of independent features and dependent features. The more the dependency the more important will be the feature. So accordingly we will be selecting the features based on the output we receive on implementing these techniques.
I will recommend you to read the blog on feature selection techniques for understanding the working of these techniques before moving to its implementation.



Feature selection python
Now comes the fun part!!!
We will see the practical implementation of this concept. I have taken a data set wine.csv.This dataset was taken purposely. This dataset contains no missing, NaN values and categorical features. This simply means the data preprocessing step is not much required.
Problem statement: Chemical analysis of wines grown in the same region and to determine the quantities of 13 constituents found in each of the three types of wines.
Pearson Correlation
Importing libraries and reading dataset
import pandas as pd import numpy as np import seaborn as sns import matplotlib.pyplot as plt %matplotlib inline df = pd.read_csv("wine.csv") df
If you are new to machine learning coding let me tell you that you have to load the dataset first before executing the above code.
Independent and dependent variables
y = data['Class'] #Load X Variables into a Pandas Dataframe with columns X = data.drop(['Class'], axis = 1) y is dependent variable and X is independent variable.
Split the data into train and test data
from sklearn.model_selection import train_test_split X_train, X_test, y_train, y_test = train_test_split(X, y,random_state=100 test_size=0.3)
Using Pearson Correlation
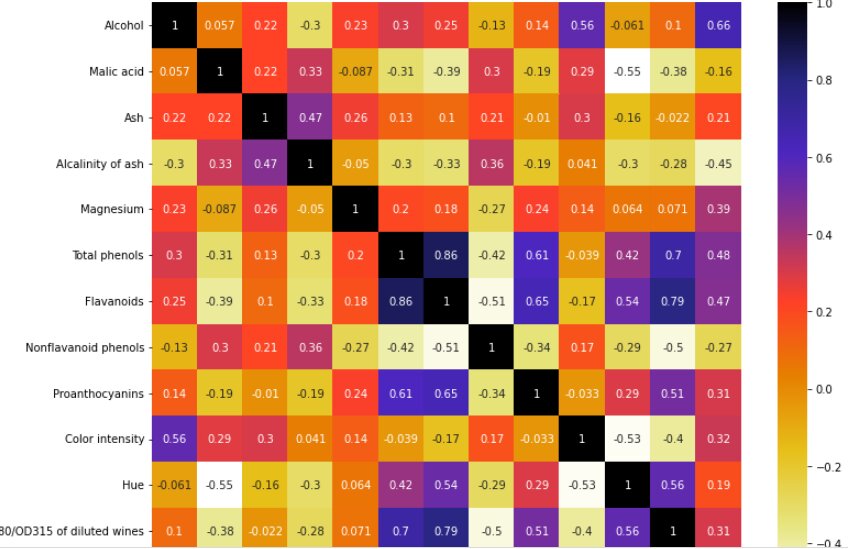
cor = X_train.corr() plt.figure(figsize=(12,10)) sns.heatmap(cor, cmap=plt.cm.CMRmap_r,annot=True) plt.show()
This heatmap shows the correlation of all the features of the dataset.
NOTE: If the value is near to 1 that means those two features are correlated and we can drop any one of them.
Finding the correlated features
def correlation(dataset, threshold): col_corr = set() corr_matrix = dataset.corr() for i in range(len(corr_matrix.columns)): for j in range(i): if abs(corr_matrix.iloc[i, j]) > threshold: colname = corr_matrix.columns[i] col_corr.add(colname) return col_corr
With the help of this function, we can select highly correlated features. The purpose is to select the correlated features and then remove them. If you note then we have written abs means absolute, because we will get a negative correlation also which will convert into a positive.
Now the very important question arises here Can we do the same for test data also. The answer is NO. Why? In order to avoid overfitting. Here we have to set the threshold value for correlation also.
Getting correlated features
corr_features = correlation(X_train, 0.7) Corr_features Output: {'Flavanoids', 'OD280/OD315 of diluted wines'}
Like here we set threshold=0.7 which means the features which are 70% or more correlated will be returned in the output. You can set any value of the threshold. Most people set it to be 0.85.
Removing correlated features
X_train.drop(corr_features,axis=1) X_test.drop(corr_features,axis=1) X_train
Chi-square
Perform chi2 test
from sklearn.feature_selection import chi2 Calculating Fscore and p value f_p_values=chi2(X_train,y_train) F_p_values output: (array([4.22853570e+00, 2.37537819e+01, 6.85971917e-01, 2.51905256e+01, 2.98617377e+01, 1.30515334e+01, 5.15232527e+01, 1.51782774e+00, 8.08914504e+00, 8.94909592e+01, 4.33089278e+00, 2.02933327e+01, 1.33574764e+04]), array([1.20721644e-01, 6.94915188e-06, 7.09648176e-01, 3.38802707e-06, 3.27797807e-07, 1.46519534e-03, 6.48437052e-12, 4.68174650e-01, 1.75171913e-02, 3.69219925e-20, 1.14698723e-01, 3.92065660e-05, 0.00000000e+00]))
Chi-square is used to find F-score and p-values for all features. So in this case the first array is for F score and the second array is for p- values. Now, what is the importance of these two values? And the more the value of the F score the more important the feature is and the lesser the value of the p-value the more important will be the feature.
Representing in list form
import pandas as pd p_values=pd.Series(f_p_values[1]) p_values.index=X_train.columns P_values
Sorting values in ascending order
p_values.sort_index(ascending=False) Output: Total phenols 1.465195e-03 Proline 0.000000e+00 Proanthocyanins 1.751719e-02 OD280/OD315 of diluted wines 3.920657e-05 Nonflavanoid phenols 4.681746e-01 Malic acid 6.949152e-06 Magnesium 3.277978e-07 Hue 1.146987e-01 Flavanoids 6.484371e-12 Color intensity 3.692199e-20 Ash 7.096482e-01 Alcohol 1.207216e-01 Alcalinity of ash 3.388027e-06 dtype: float64
Observation
The total phenols Column is the most important column when compared to the output feature.
If we have written ascending=True then features would be in ascending order.
Now let’s learn the third very important technique i.e feature selection by using mutual information gain.
Mutual information gain
Importing mutual information gain
from sklearn.feature_selection import mutual_info_classif # determine the mutual information mutual_info = mutual_info_classif(X_train, y_train) mutual_info Output: array([0.45810181, 0.26490518, 0.05332173, 0.2187717 , 0.25110079, 0.41649437, 0.68865959, 0.08089527, 0.29778143, 0.5377321 , 0.47179421, 0.48180695, 0.53742616])
mutual_info_classif is a library that is present in feature_selection. It is the property of mutual information gain that the value you will get will never be negative. The higher the value the more important that feature will be or you can say that the dependency of that independent feature will be more on the dependent feature.
Representing in list form
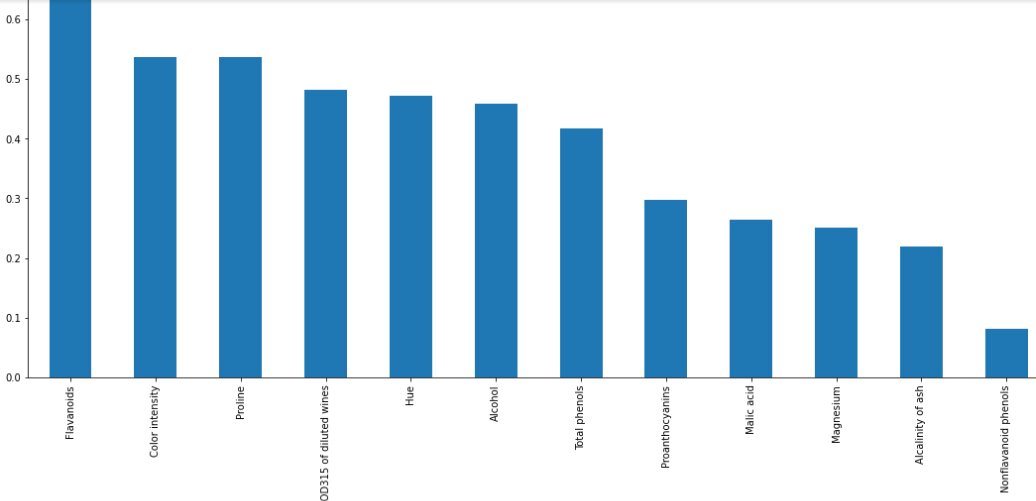
mutual_info = pd.Series(mutual_info) mutual_info.index = X_train.columns mutual_info.sort_values(ascending=False) Output: Flavanoids 0.688660 Color intensity 0.537732 Proline 0.537426 OD280/OD315 of diluted wines 0.481807 Hue 0.471794 Alcohol 0.458102 Total phenols 0.416494 Proanthocyanins 0.297781 Malic acid 0.264905 Magnesium 0.251101 Alcalinity of ash 0.218772 Nonflavanoid phenols 0.080895 Ash 0.053322 dtype: float64
Plotting the graph
plot the ordered mutual_info values per feature
mutual_info.sort_values(ascending=False).plot.bar(figsize=(20, 8)
Selecting best 5 features
from sklearn.feature_selection import SelectKBest #No we Will select the top 5 important features sel_five_cols = SelectKBest(mutual_info_classif, k=5) sel_five_cols.fit(X_train, y_train) X_train.columns[sel_five_cols.get_support()] Output: Index(['Flavanoids', 'Color intensity', 'Hue', 'OD280/OD315 of diluted wines','Proline'],dtype='object')
Best-suited Machine Learning courses for you
Learn Machine Learning with these high-rated online courses
Assignment
I suggest you download this dataset and try these techniques first and then try it on some big datasets. You will get the same dataset on Kaggle and Github. You can try the Pima Indians Diabetes dataset also for your practice.
Endnotes
We can judge features’ importance by ourselves also, based on our knowledge and experience. This blog included the code for the different feature selection techniques.
If this blog helped you in any way then please share it with other data science aspirants also. If you are interested in going into the data science field then you find different study material on this page.
Share knowledge!!!
Happy Learning!!!
This is a collection of insightful articles from domain experts in the fields of Cloud Computing, DevOps, AWS, Data Science, Machine Learning, AI, and Natural Language Processing. The range of topics caters to upski... Read Full Bio